# Table of Content
# Abbreviations
ML Machine Learning
DL Deep Learning
CNN Convolutional Neural Network
DCNN Deep Convolutional Neural Network
ILSVRC ImageNet Large Scale Visual Recognition Challenge
VGG Visual Geometry Group ( Convolutional neural network architecture )
COCO Common Objects in Context
# Abstract
With the increase in population, the needs have also been increased. As agriculture is a vital source for providing food, whereas crop ailments are major commination to agriculture, and their early detection remains strenuous over the globe due to insufficient technology, agricultural organizations are unable to reach farmers in time for necessary precautionary measures. As a result, farmers have to suffer from compromised lower crop yield. Many machine learning models were used to detect and identify diseases of plants but, after the advancement in Deep Learning, this field seems to have great potential concerning improved accuracy. The combination of advancements in computer vision and the global smartphone penetration made possible by deep learning to provide a mobile-phone-based system to diagnose diseases. Using a public data-set of 87,900 photos of healthy and diseased leaves, several models were trained to identify 14 crops and the presence or absence of 26 diseases, with the best performance reaching a 98.58% on the retained dataset, which demonstrates the practicality of our perspective. Generally, the methodology of training deep convolutional neural networks on exceptionally huge and publicly accessible image data-sets presents a straightforward path towards a massive global diagnosis of mobile-phone-assisted crop disease.
# Chapter 1: Introduction
Earth is occupying more than 7 billion people. The number is increasing gradually. With a persistent increase in population, it is understood that basic life necessities are also increasing in which food is on top of the list. The increasing population has also eaten up the land, and the area for agricultural land is shrinking up. So we have to get an adequate amount of food from the land available. Food sustainability, however, remains endanger by a variety of factors which includes climate change [5], plant diseases [23] and others. When the plants are growing up, they are attacked by diseases which clearly means a compromised lower crop yield. Unfortunately, smallholder farmers have to face the disastrous consequences whose income entirely depends upon crops. Yield. The major agricultural production comes from smallholder farmers [9], and they have to suffer approximately 50% yield loss due to climate change, pest attack, and diseases. Acknowledging these problems, various attempts have been made to avert or lower the loss of crop yield. To prevent the disease, it is crucial to detect the disease at an early stage. And efficient disease management is a very crucial step in this regard. Agricultural organizations have been working for disease detection at an early stage at local clinics. During the past decades the world has totally turned into ”Global Village” and because of that enormous data is available online including information on disease diagnosis [11] and the leverage of which is internet penetration worldwide. More recently, mobile phone technology incredibly has become famous due to the proliferation of mobile-based tools in all parts of the world. All these factors escort us to a point where disease detection is technically feasible and available at an unparalleled scale. Unlike other countries, Pakistan lacks modern technology due to which, we are unable to detect diseases in time and to reach farmers in order to raise awareness about rehabilitation. The blue-collar approaches make it very slow for policy-making organizations to gather data and draw results for immediate movement. The intensity behind this research work is to dispense an efficient system that can detect the disease straight-away whether a farmer or agricultural organization uses it. We intended to develop mobile as well as a web-based tool using deep learning for the detection of crop diseases. Deep learning has proved its worth successfully in many different domains such as end-to-end learning. Now we are going to demonstrate the technical feasibility of our proposed approach by utilizing 87,900 images on healthy and infected leaves of crop plants that are openly available on the online system PlantVillage [2].

Figure 1.1: Sample leaf images from the dataset
A DCNN includes the mapping between an input to an output. Deep learning is probability-based means it never gives us the definite answer however it gives us the probabilities. The term CNN itself stipulates that a mathematical function called convolution is used within the network. Convolution is a specific type mathematical operation on two functions which produces a third function that expresses how another changes one’s form. A CNN consists of two main input and output layers, and multiple hidden layers. Typically the hidden layers consist of a series of convolutionary layers. The activation function is commonly a Rectified Linear Units (RELU). The purpose of RELU is to normalize the values after the application of convolution to convert the values in a specific range. Additional convolutions such as pooling, fully connected layers and normalization layers follow the RELU. Pooling is to choose the best or one thing out of the pool of things. The nodes in neural networks are computational units which take weighted inputs from the incoming edges and provide an outgoing edge with numerical output. node enumerates an output value by adding a specific function to the previous layer’s input values. In a neural network, model learning progresses through iterative changes to these weights and biases. DCNN is learned by changing network parameters in such a way as to improve mapping during the training. For thepurpose of plant disease identification, we needed a large and verified data-set of healthy and diseased images to train an accurate image classifier, but such dataset did not exist until very recently, and even small dataset were not publicly available. In order to tackle this issue, the PlantVillage project began to collect a large dataset of diseased and healthy crop plants and made them available publicly. We announce here on the classification of 26 diseases (presence or absence) in 14 crop species using 87,900 with deep learning (DL).
# Chapter 2: Motivation and Problem Statement
# 2.1: Motivation
With the increase in population, the needs have also been increased. As agriculture is the vital source for providing food but, unfortunately, because of lack the modern technology, the agricultural organization are unable to reach farmers in time for necessary precautionary measures. As a results farmers have to suffer from compromised lower crop yield. We are aimed to work for a solution that can prevent farmers from suffering a loss to some extent.
# 2.2: Problem Statement
Plant disease affects not only the production of human food but also natural systems. The majority of smallholder farmers do not have access to the resources that can identify the diseases accurately and timely. Due to the lack of timely and accurate agricultural information, they have to suffer from lower crop yield.
# Chapter 3: Literature Review
The Deep Learning (DL) is subcategory of ML. This field is still evolving. Machine Learning has made enormous evolution in the past few years. Many advances were found in the first phase, such as handwritten text recognition, back-propagation and resolving training problems. In the second phase was to develop algorithms for health-sectors, text-recognition, earthquake-predictions, marketing, finance, image-recognition, and object detection. In 2012 a deep convolutionary neural network accomplished a top-5 error of 15.3% when classifying images into 1000 possible categories [1]. In the next three years, numerous advances in convolutionary neural networks reduced the error rate to 3.57. With the passage of time as Deep Learning architectures started to evolve, researchers applied them to classification, segmentation, object detection, video processing, natural language processing, image recognition, and speech recognition. Different agriculture application has also been developed using these architectures. For example, Leaf counting was performed using Deep CNN with average accuracy of 95% [25]. Leaf classification was performed using deep convolutional neural network classifier among 32 different species with average accuracy of 97.3% [7]. Fruit counting was performed by using simulated deep convolutional neural network with an average accuracy of 91% [19]. Classification of land cover and crop type was performed by using deep learning classifier with accuracy of crop type identification of 95% [18]. Identification of plants was performed by using Deep convolutional neural network [10], [16]. In [26] identification of plants was performed among 100 different species utilizing 10,000 images using deep learning with an accuracy of 91.78%. In addition deep learning techniques are also used in for crucial tasks such as crop plant disease identification and classification which is main topic of this thesis. For example, in [20] plant disease detection was performed using deep convolutional neural network classifier. To sum up, used DL Architectures are shown in the table along with the selected plants and their results.
| Deep Learning Techniques | Dataset | Used Plants | Accuracy | Reference |
|---|---|---|---|---|
| Convolutional Neural Network | Plant Village | Maize | 92.85% | [21] |
| LeNet | Plant Village | Banana | 98.54% | [4] |
| Alex-Net, VGG16, VGG-19, Squeeze-Net, Goog-LeNet, Inception-v-3, Inception-ResNet-v-2, ResNet-50, Resnet-101 | Real Field Dataset | Apricot, Walnut, Peach, Cherry | 97.14% | [24] |
| Inception-v-3 | Experimental Field Dataset | Cassava | 93% | [3] |
| Convolutional Neural Network | Images Taken From The Research Center | Cucumber | 82.3% | [8] |
| Super Resolution Convolutional Neural Network (SCRNN) | Plant Village | Tomato | 90% | [15] |
| Caffe-Net | Downloaded From The Internet | Pear, cherry, peach, apple, grapevine | 96.3% | [22] |
| Resnet-50, Inception-V-2, Mobile-Net-V-1 | Real Environment | Banana | 99% of ResNet-50 | [17] |
| Mobile-Net, Modified-Mobile-Net, Reduced-Mobile-Net | Plant Village dataset | 24 Types Of Plants | 98.34% of reduced MobileNet | [14] |
Table 3.1: DL Architectures along with selected plant species and results
From this table, we can conclude that while some Deep Learning Architectures/- Models have been developed for the identification of the diseases of plants but this is still a prolific research field and should lead to improvements for better plant disease identification. So we needed a large and verified data set of healthy and diseased images to train an accurate image classifier for plant disease diagnosis, but such a dataset did not exist until very recently. Plant Village collected a huge dataset of 87,900 images of plant health to enable the development of smartphone assisted disease diagnosis and made it publicly and freely available [12].
# Chapter 4: Materials and Methods
# 4.1: Materials
Our dataset is consists of 87,900 pictures of healthy and diseased plant leaves. All the images in the dataset were taken at experimental research stations affiliated with Land Grant Universities in the USA (Penn State, Florida State, Cornell, and others) [12]. These images consist of 26 major crop diseases (4 bacterial diseases, 17 Fungal diseases, 2 viral diseases, 2 mold diseases, and 1 disease caused by a mite). These images also have 12 healthy leaves of 12 crop species that are not visibly affected by a disease. Table 4.1 Summarizes the dataset.
| Crop | Classes |
|---|---|
| Apple | Apple-Healthy, Apple-Cedar-Rust, Apple-Black-Rot, Apple-Scab |
| Raspberry | Raspberry-Healthy |
| Soybean | Soybean-Healthy |
| Squash | Squash-Powdery-Mildew |
| Strawberry | Strawberry-Leaf-Scorch, Strawberry-Healthy |
| Tomato | Tomato-Healthy, Tomato-Late-Blight, TomatoBacterial-Spot, Tomato-Early-Blight, TomatoLeaf-Mold, Tomato-Septoria-Leaf-Spot, TomatoMosaic-Virus, Tomato-Two-Spotted-Spider-Mite, Tomato-Target-Spot, Tomato-Yellow-Leaf-CurlVirus |
| Potato | Potato-Early-Blight, Potato-Late-Blight, PotatoHealthy |
| Blueberry | Blueberry-Healthy |
| Cherry | Cherry-Healthy, Cherry-Powdery-Mildew |
| Corn | Corn-Healthy, Corn-Common-Rust, Corn-Gray-Leaf Spot, Corn-Northern-Leaf-Blight |
| Grape | Grape-Healthy, Grape-Leaf-Blight, Grape-BlackRot, Grape-Black-Measles(Esca) |
| Orange | Orange-Huanglongbing (Citrus-Greening) |
| Peach | Peach-Healthy, Peach-Bacterial-Spot |
| Bell-Pepper | Bell-Pepper-Bacterial-Spot, Bell-Pepper-Healthy |
Table 4.1: List of Crops with their respective classes in the Dataset.
# 4.2: Methods
Since these diseases can severely affect plants, we landed up with four different models for 38 different classes (Table 4.1) to achieve maximum accuracy. Blueprint of our proposed system for disease detection is showed in the figure 4.1.
To train the model, we focused on 4 different architectures, namely VGG16, Resnet152, MobileNet, and MobileNetV2.
VGG16 was proposed by Andrew Zisserman and Karen Simonyan of the Visual Geometry Community Lab at Oxford University. This model secured 1st and 2nd place in ImageNet Large Scale Visual Recognition Challenge (ILSVRC) in 2014. The VGG16 network is trained on an ImageNet dataset which has 14 million images and 1000 classes, and this model achieves 92.7% top-5 test accuracy.
Resnet was proposed by Microsoft Research Asia [13] and won the ImageNet Large Scale Visual Recognition Challenge (ILSVRC) and MS-COCO competition in 2015. ResNet architecture has several variants all of them have the similar concept but have different number of layers, for example ResNet-18, ResNet-50, ResNet-101 and so on. The name Resnet followed by numbers simply insinuate the Resnet architecture witch has number of layers of neural network. The main idea manoeuvred in these models, residual network connections, is found to greatly enhance gradient flow, thus enabling the training of tens or even hundreds of layers of much deeper models.
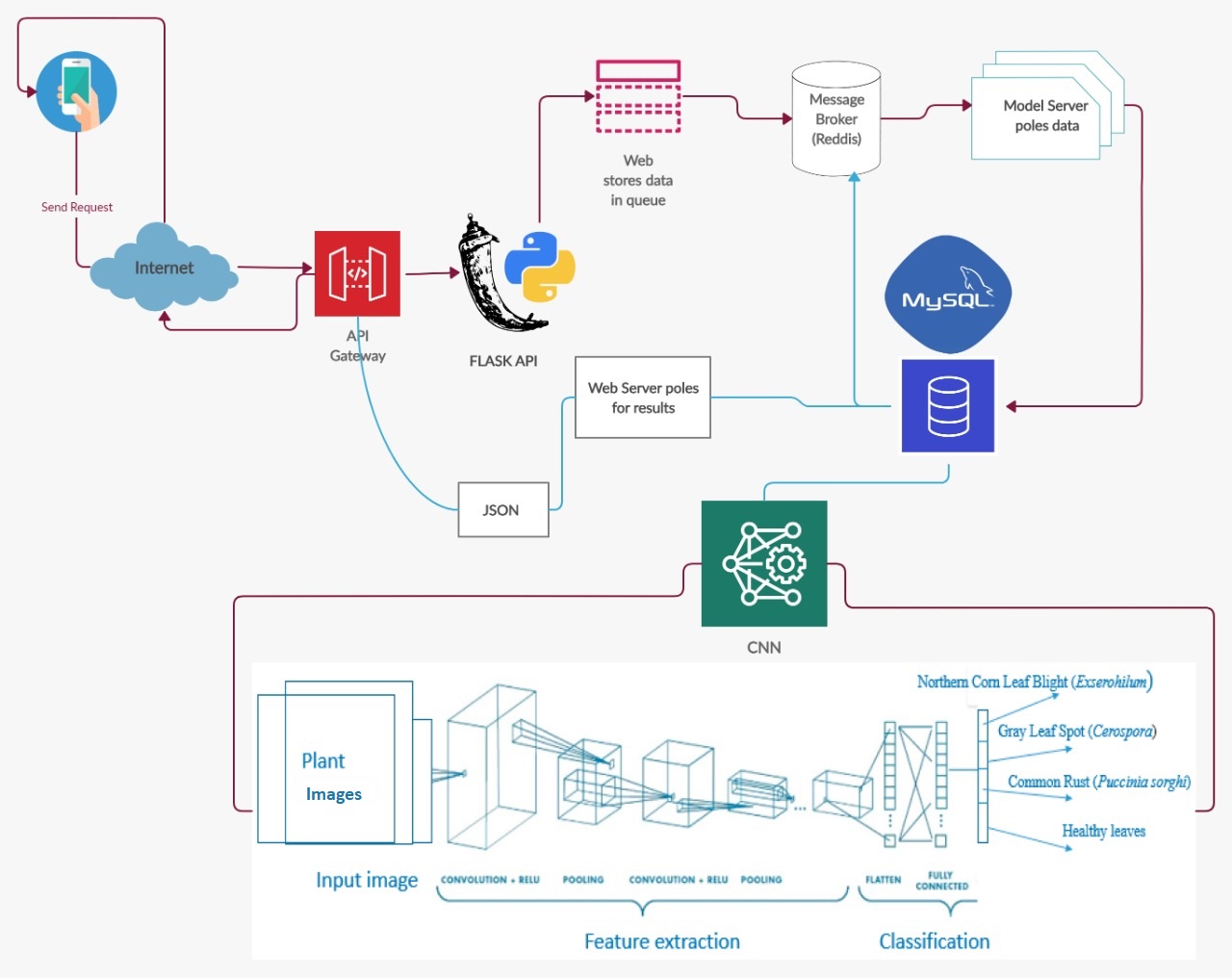
Figure 4.1: Blueprint of our proposed system for disease detection
MobileNet is a model built primarily from depthwise separable convolutions to create lightweight deep convolutional neural networks and provides an efficient model for smartphone and integrated vision applications [6].
To train these models we used Google Colaboratory framework. Google Colab is a free cloud service created by Google, in which the person who has a Gmail account can write, execute codes for Machine learning as well as for Deep learning. In Google Colab different runtime environments and Various versions of python are available. It can also download large datasets to google drive at higher speed directly from the servers. With Google Colab, you can also mount your drive and it can fetch the appropriate file after the authentication. The most important feature of Google Colab that distinguishes it from other free cloud services is it provides GPU and it is totally free. To summarize, we have 4 experimental configurations in total, which are described in the next section
# 4.2.1: Experiment # 1
In this experiment, we used a python deep learning library called PyTorch, and the model we used in this experiment is Resnet152. We used transfer learning to train the model because it always yields better results. This experiment runs for a total of 50 epochs and achieves testing accuracy of 98.5%.
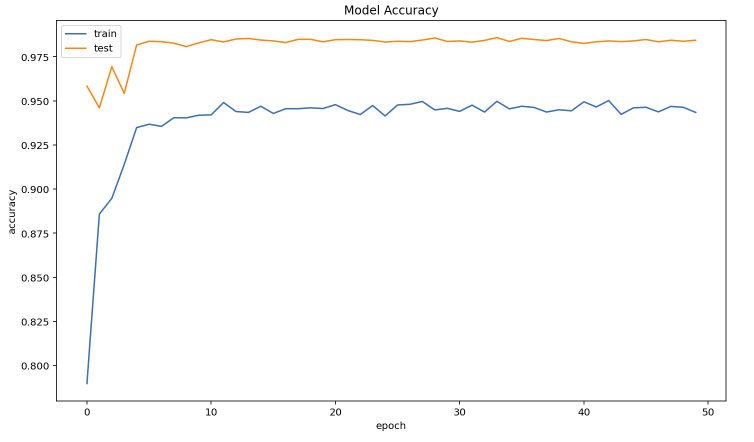
Figure 4.2: Model Accuracy
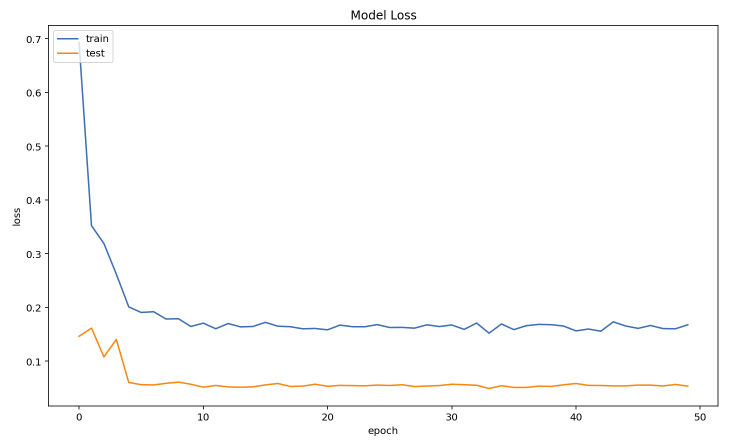
Figure 4.3: Model Loss
# 4.2.2: Experiment # 2
In this experiment, we used a python deep learning library called TensorFlow, and we created a custom model. This experiment runs for a total of 80 epochs and achieves an accuracy of 94.72%.

Figure 4.4: Accuracy of the classifier for all classes
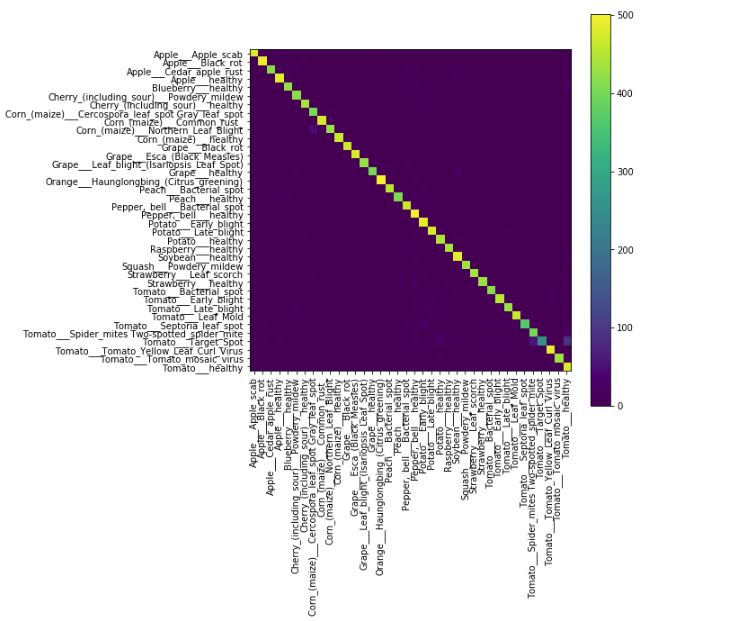
Figure 4.4: Graph of all Classes and their Identifications
# Chapter 5: Implementation Details
The very first was to gather the data and train the model to get the results. After the collection of the dataset which is consisted of 87,900 different plant images, we divided the dataset into train-test splits of 25-75, 50-50, and 25-75. We made these splits to get a sense of how our proposed approach will perform on the unseen data and also keep track of if our proposed approach is underfitting or overfitting. After training the model with the best performance reaching a 98.58% on a held-out test set we developed an API using Flask for this model to interact with our mobile application. After the development of the API, we developed a mobile application to interact with the API which interacts with the model to give the final results.
# 5.1: Snippets of Model Training Code
Here are some snippets of our code to train the model.
import torch
import numpy as np
import matplotlib.pyplot as plt
import torch
import time
import numpy as np
from torch import nn, optim
import torch.nn.functional as F
from torchvision import datasets, transforms, models
import torchvision
from collections import OrderedDict
from torch.autograd import Variable
from PIL import Image
from torch.optim import lr_scheduler
Figure 5.1: Libraries used in the model training
data_dir = '/content/DataSet'
train_dir = data_dir + '/train'
valid_dir = data_dir + '/val'
nThreads = 4
batch_size = 32
Figure 5.2: Load the dataset
data_transforms = {
'train': transforms.Compose([
transforms.RandomRotation(30),
transforms.RandomResizedCrop(224),
transforms.RandomHorizontalFlip(),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
]),
'val': transforms.Compose([
transforms.Resize(256),
transforms.CenterCrop(224),
transforms.ToTensor(),
transforms.Normalize([0.485, 0.456, 0.406], [0.229, 0.224, 0.225])
]),
}
data_dir = '/content/DataSet'
image_datasets = {x: datasets.ImageFolder(os.path.join(data_dir, x),
data_transforms[x])
for x in ['train', 'val']}
dataloaders = {x: torch.utils.data.DataLoader(image_datasets[x], batch_size=batch_size,
shuffle=True, num_workers=4)
for x in ['train', 'val']}
dataset_sizes = {x: len(image_datasets[x]) for x in ['train', 'val']}
class_names = image_datasets['train'].classes
Figure 5.3: Applying Data Augmentation
model = models.resnet152(pretrained=True)
for param in model.parameters():
param.requires_grad = False
from collections import OrderedDict
classifier = nn.Sequential(OrderedDict([
('fc1', nn.Linear(2048, 512)),
('relu', nn.ReLU()),
('fc2', nn.Linear(512, 39)),
('output', nn.LogSoftmax(dim=1))
]))
model.fc = classifier
Figure 5.4: Building the Classifier
def train_model(model, criterion, optimizer, scheduler, num_epochs=20):
since = time.time()
best_model_wts = copy.deepcopy(model.state_dict())
best_acc = 0.0
for epoch in range(1, num_epochs+1):
print('Epoch {}/{}'.format(epoch, num_epochs))
print('-' * 10)
for phase in ['train', 'val']:
if phase == 'train':
scheduler.step()
model.train()
else:
model.eval()
running_loss = 0.0
running_corrects = 0
for inputs, labels in dataloaders[phase]:
inputs, labels = inputs.to(device), labels.to(device)
optimizer.zero_grad()
with torch.set_grad_enabled(phase == 'train'):
outputs = model(inputs)
loss = criterion(outputs, labels)
_, preds = torch.max(outputs, 1)
if phase == 'train':
loss.backward()
optimizer.step()
running_loss += loss.item() * inputs.size(0)
running_corrects += torch.sum(preds == labels.data)
epoch_loss = running_loss / dataset_sizes[phase]
epoch_acc = running_corrects.double() / dataset_sizes[phase]
print('{} Loss: {:.4f} Acc: {:.4f}'.format(
phase, epoch_loss, epoch_acc))
if phase == 'val' and epoch_acc > best_acc:
best_acc = epoch_acc
best_model_wts = copy.deepcopy(model.state_dict())
print()
time_elapsed = time.time() - since
print('Training complete in {:.0f}m {:.0f}s'.format(
time_elapsed // 60, time_elapsed % 60))
print('Best valid accuracy: {:4f}'.format(best_acc))
model.load_state_dict(best_model_wts)
return model
Figure 5.5: Function to train the model
num_epochs = 80
criterion = nn.NLLLoss()
optimizer = optim.Adam(model.fc.parameters(), lr=0.001)
exp_lr_scheduler = lr_scheduler.StepLR(optimizer, step_size=5, gamma=0.1)
model_ft = train_model(model, criterion, optimizer, exp_lr_scheduler, num_epochs=num_epochs)
Figure 5.6: : Start Training
def test(model, dataloaders, device):
model.eval()
accuracy = 0
model.to(device)
for images, labels in dataloaders['val']:
images = Variable(images)
labels = Variable(labels)
images, labels = images.to(device), labels.to(device)
output = model.forward(images)
ps = torch.exp(output)
equality = (labels.data == ps.max(1)[1])
accuracy += equality.type_as(torch.FloatTensor()).mean()
print("Testing Accuracy: {:.3f}".format(accuracy/len(dataloaders['val'])))
test(model, dataloaders, device)
Figure 5.7: Model validation after Training
model.class_to_idx = dataloaders['train'].dataset.class_to_idx
model.epochs = num_epochs
checkpoint = {'input_size': [3, 224, 224],
'batch_size': dataloaders['train'].batch_size,
'output_size': 39,
'state_dict': model.state_dict(),
'data_transforms': data_transforms,
'optimizer_dict':optimizer.state_dict(),
'class_to_idx': model.class_to_idx,
'epoch': model.epochs}
torch.save(checkpoint, 'plants9615_checkpoint.pth')
Figure 5.8: Saving the model for Further use
# 5.2: Snippets of App User Interface
User interface for the mobile application is shown in the upcoming pictures.
Figure 5.9: Sign Up Screen
Figure 5.10: Login Screen

Figure 5.11: Home Screen
Figure 5.12: Community Screen
Figure 5.13:User Screen
Figure 5.14:Add Post Screen

Figure 5.15: Logout Screen
Figure 5.16: Prediction # 1 Screen

Figure 5.17: Prediction # 2 Screen

Figure 5.18: Prediction # 3 Screen
Figure 5.19: Prediction # 4 Screen

Figure 5.20: Prediction # 5 Screen
# Chapter 6: Results
All results were observed on the premise that the trained model has to identify the crop genus and the state of the infection. After training model with different configurations the overall accuracy we procured on the collected dataset varied from 75% to 98.58% which showing us the strong promise of a DL approach to similar problems. Mean accuracy’s across our experiments is shown in the table given below.
| Experiment | Category | Dataset Splits | Model Used | Epochs | Mean Accuracy |
|---|---|---|---|---|---|
| 1 | Transfer Learning | 70% Train, 30% Validation, Testing | VGG16 | 30 | 94.72% |
| 2 | Transfer Learning | 70% Train, 30% Validation, Testing | Resnet152 | 30 | 96.43% |
| 3 | Transfer Learning | 70% Train, 30% Validation, Testing | Resnet152 | 50 | 98.5849% |
| 4 | Custom Model | 70% Train, 30% Validation, Testing | Created Custom Model | 50 | 96% |
Table 6.1: Mean accuracy’s across all experiments
# References
[1] Ilya Sutskever Alex Krizhevsky and Geoffrey E. Hinton. ImageNet Classification with Deep Convolutional Neural Networks. Online Available at: https://papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks.pdf
[2] Abdallah Ali. PlantVillage Dataset Dataset of diseased plant leaf images and corresponding labels. Available at: https://www.kaggle.com/xabdallahali/plantvillage-dataset
[3] Peter McCloskey Babuali Ahmed Ally James Legg Amanda Ramcharan, Kelsee Baranowski and David P. Hughes. Deep Learning for Image-Based Cassava Disease Detection. Available at: . https://www.frontiersin.org/articles/10.3389/fpls.2017.01852/full?report=reader.
[4] Bouaziz Bassem Amara Jihen and Algergawy Alsayed. A Deep Learningbased Approach for Banana Leaf Diseases Classification. Available at: . https://dl.gi.de/handle/20.500.12116/944.
[5] Maria Val Martin Amos P. K. Tai and Colette L. Heald. Threat to future global food security from climate change and ozone air pollution. Available online at. https://www.nature.com/articles/nclimate2317.
[6] Bo Chen Dmitry Kalenichenko Weijun Wang Tobias Weyand Marco Andreetto Andrew G. Howard, Menglong Zhu and Hartwig Adam. MobileNets: Efficient Convolutional Neural Networks for Mobile Vision Applications. Available at: . https://arxiv.org/pdf/1704.04861.pdf.
[7] Feras Dayoub Niko Sunderhauf David Hall, Chris McCool and Ben Upcroft. Evaluation of Features for Leaf Classification in Challenging Conditions. Available at: . https://ieeexplore.ieee.org/abstract/document/7045965.
[8] Hiroyuki Uga Satoshi Kagiwada Erika Fujita, Yusuke Kawasaki and Hitoshi Iyatomi. Basic Investigation on a Robust and Practical Plant Diagnostic System. Available at: . https://ieeexplore.ieee.org/abstract/document/7838282.
[9] International Fund for Agricultural Development (IFAD) andUnited Nations Environment Programme (UNEP). Smallholders, food security, and the environment. Available online at. https://www.ifad.org/documents/38714170/39135645/smallholders_report.pdf/133e8903-0204-4e7d-a780-bca847933f2e.
[10] Monica G.Larese Guillermo L.Grinblat, Lucas C.Uzal and Pablo M.Granitto. Deep learning for plant identification using vein morphological patterns. Available at: . https://www.sciencedirect.com/science/article/pii/S0168169916304665.
[11] David. P. Hughes and Marcel Salathe. An open access repository of images on plant health to enable the development of mobile disease diagnostics. Available online at. https://arxiv.org/abs/1511.08060
[12] David P. Hughes and Marcel Salathe. An open access repository of images on plant health to enable the development of mobile disease diagnostics. Available at: . https://arxiv.org/ftp/arxiv/papers/1511/1511.08060.pdf
[13] Shaoqing Ren Jian Sun (Microsoft Research) Kaiming He, Xiangyu Zhang. Deep Residual Learning for Image Recognition. Available at: .https://arxiv.org/pdf/1512.03385.pdf.
[14] Mingyang Wu Kamal KC, Zhendong Yin and Zhilo Wu. Depthwise separable convolution architectures for plant disease classification.Available at: . https://www.sciencedirect.com/science/article/pii/S0168169918318696.
[15] Takashi Togami Kyosuke Yamamoto and Norio Yamaguchi. Super-Resolution of Plant Disease Images for the Acceleration of Image-based Phenotyping and Vigor Diagnosis in Agriculture. Available at: . https://www.mdpi.com/1424-8220/17/11/2557.
[16] Sue Han Lee and Chee Seng Chan. Deep-plant: Plant identification with convolutional neural networks. Available at: . https://ieeexplore.ieee.org/abstract/document/7350839.
[17] Henry Ruiz Nancy Safari Sivalingam Elayabalan Walter Ocimati Michael Gomez Selvaraj, Alejandro Vergara and Guy Blomme. AI-powered banana diseases and pest detection. Available at: . https://link.springer.com/article/10.1186/s13007-019-0475-z.
[18] S. Skakun Nataliia Kussul, Mykola Lavreniuk and Andrii Shelestov. Deep Learning Classification of Land Cover and Crop Types Using Remote Sensing Data. Available at: . https://ieeexplore.ieee.org/document/7891032.
[19] Maryam Rahnemoonfar and Clay Sheppard. Deep Count: Fruit Counting Based on Deep Simulated Learning. Available at: . https://www.mdpi.com/1424-8220/17/4/905.
[20] David Hughes Sharada Prasanna Mohanty and Marcel Salathe. Using Deep Learning for Image-Based Plant Disease Detection. Available at: . https://www.frontiersin.org/articles/10.3389/fpls.2016.01419/full.
[21] Malusi Sibiya and Mbuyu Sumbwanyambe. A Computational Procedure for the Recognition and Classification of Maize Leaf Diseases Out of Healthy Leaves Using Convolutional Neural Networks. Available at: . https://www.mdpi.com/2624-7402/1/1/9.
[22] Andras Anderla Dubravko Culibrk Srdjan Sladojevic, Marko Arsenovic and Darko Stefanovic. Deep Neural Networks Based Recognition of Plant Diseases by Leaf Image Classification. Available at: . https://www.hindawi.com/journals/cin/2016/3289801/.
[23] Richard N. Strange. and Peter R. Scott. Plant Disease: A Threat to Global Food Security. Available online at. https://www.annualreviews.org/doi/full/10.1146/annurev.phyto.43.113004.133839.
[24] MUAMMER TURKOGLU and DAVUT HANBAY. Plant disease and pest detection using deep learning-based features. Available at: . https://journals.tubitak.gov.tr/elektrik/abstract.htm?id=24806.
[25] Faina Khoroshevsky Alon Shpigler Aharon Bar Hillel Yotam Itzhaky, Guy Farjon. Leaf Counting: Multiple Scale Regression and Detection Using Deep CNNs. Available at: . https://www.plant-phenotyping.org/lw_resource/datapool/systemfiles/elements/files/63b7a231-949c-11e8-8a88-dead53a91d31/current/document/0031.pdf.
[26] Guan Wang Yu Sun, Yuan Liu and Haiyan Zhang. Deep Learning for Plant Identification in Natural Environment. Available at: . https://www.hindawi.com/journals/cin/2017/7361042/.